Published online Feb 28, 2022. doi: 10.3748/wjg.v28.i8.811
Peer-review started: October 1, 2021
First decision: November 7, 2021
Revised: November 19, 2021
Accepted: January 14, 2022
Article in press: January 14, 2022
Published online: February 28, 2022
Nodular lymphoid hyperplasia (NLH) in the small intestine is a rare benign lesion characterized by multiple small nodules on the intestinal surface. Patients with terminal ileal NLH may experience long-term abdominal pain, diarrhea, and abdominal distension, among other symptoms. Supplementation with probiotics could mitigate these symptoms. NLH is linked to the immune system, and it may result from accumulation of plasma-cell precursors due to a maturational defect during the development of B lymphocytes. The intestinal microbiome plays an essential role in the immune system. Thus, we speculate that the gut flora plays a key role in terminal ileal NLH.
To explore the correlation between intestinal flora and terminal ileal NLH.
We collected mucosal biopsy samples that were obtained via colonoscopy from 15 patients with terminal ileal NLH (the test group) and 15 normal subjects (the control group). We subsequently performed 16S-rRNA gene amplicon sequencing of these samples, and the results were evaluated using alpha diversity, beta diversity and microbial composition analyses. The Phylogenetic Investigation of Communities by Reconstruction of Unobserved States was used to predict the metabolic pathways and orthologous groups according to the Kyoto Encyclopedia of Genes and Genomes database.
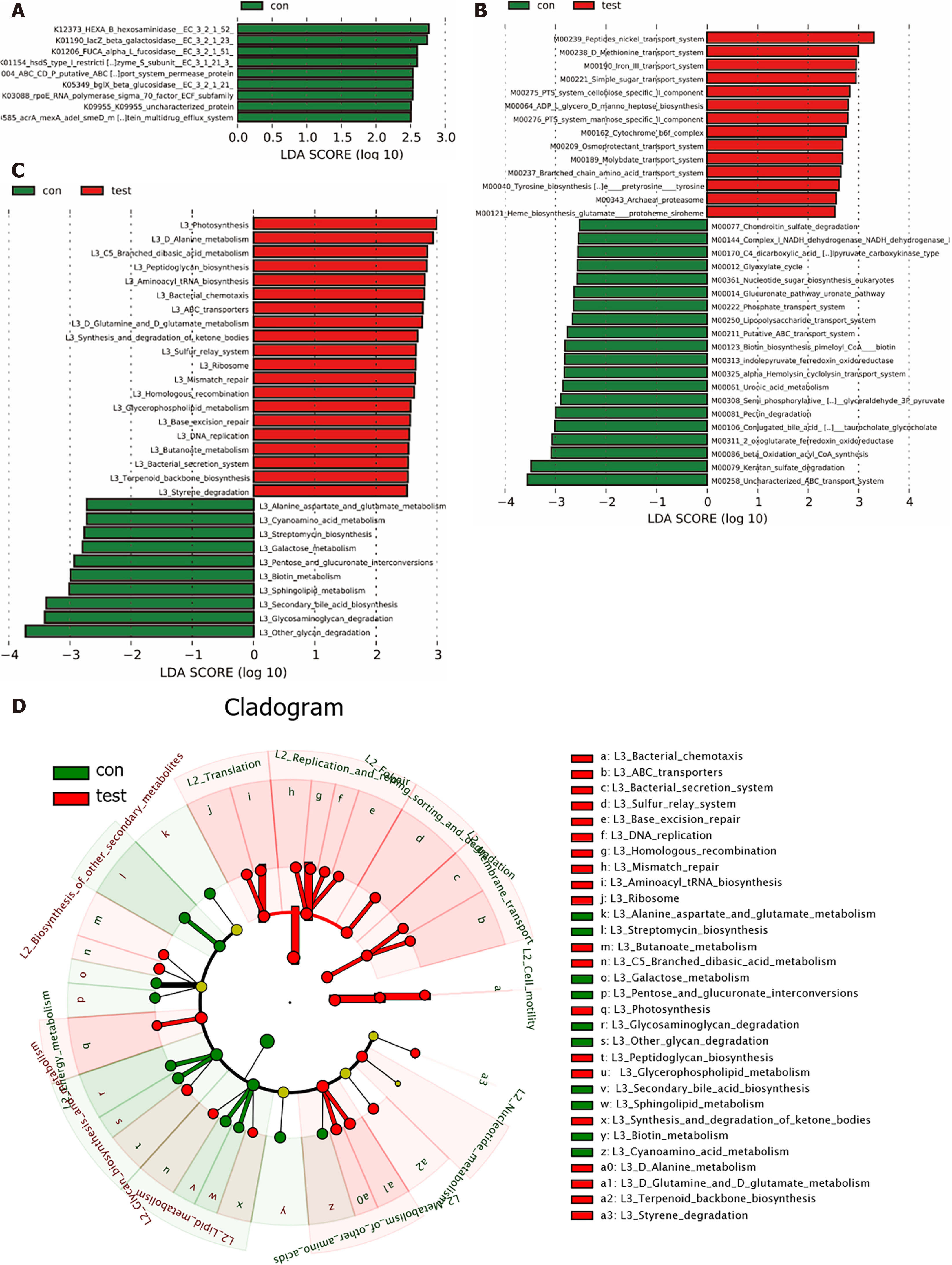
Compared with the control group, the terminal ileal NLH group showed an increased alpha diversity (P < 0.05). The overall intestinal microbiota in the NLH group was significantly different from that of the control group (P < 0.05), implying that there was the dysbiosis in the terminal ileal NLH patients. The relative abundance of phylum Bacteroidetes was significantly lower in the NLH group, while that of Patescibacteria and Campilobacterota was significantly higher. The genus Bacteroides was the dominant gut microbiota in both groups, but its abundance was significantly lower in the test group than it was in the control group. Conversely, the relative abundances of Haemophilus, Streptococcus, Pseudomonas, Actinomyces, TM7X, Fusobacterium nucleatum, Parvimonas, Granulicatella, Helicobacter, and the [Eubacterium] nodatum group were significantly higher in the test group than they were in the control group. In addition, several altered metabolic pathways, orthologous groups, and modules were found. For example, the Peptidoglycan biosynthesis and Aminoacyl tRNA biosynthesis were both increased in the test group.
Maintaining the microbial balance and supplementing targeted protective bacteria could improve symptoms and potentially reduce the risk of lymphoma transformation in patients with terminal ileal NLH.
Core Tip: Nodular lymphoid hyperplasia (NLH) in the small intestine is a rare benign lesion characterized by multiple small nodules on the surface of the intestine. To explore the correlation between the intestinal flora and terminal ileal NLH, we performed bacterial 16S rRNA gene sequencing of mucosal samples from patients with terminal ileal NLH. Our results reveal that specific microflora may act on the mucosa of the small intestine and cause terminal ileal NLH. Therefore, maintaining the balance of intestinal flora and supplementing targeted protective bacteria may improve terminal ileal NLH symptoms and potentially reduce the risk of lymphoma transformation.
- Citation: Jiang QL, Lu Y, Zhang MJ, Cui ZY, Pei ZM, Li WH, Lu LG, Wang JJ, Lu YY. Mucosal bacterial dysbiosis in patients with nodular lymphoid hyperplasia in the terminal ileum. World J Gastroenterol 2022; 28(8): 811-824
- URL: https://www.wjgnet.com/1007-9327/full/v28/i8/811.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i8.811
Nodular lymphoid hyperplasia (NLH) in the small intestine is a rare benign lesion, and its incidence has not yet been determined. It is characterized by multiple small nodules on the surface of the intestine, which are found to be present in the lamina propria and superficial submucosa of the intestine[1]. The diagnosis of NLH is mainly based on endoscopic and histological examinations, markedly including the presence of hyperplastic lymphoid follicles and mitotically active germinal centers with well-defined lymphocytic mantles[2]. Terminal ileal NLH is a type of NLH, and patients with terminal ileal NLH demonstrate multiple symptoms that seriously reduce the individual’s quality of life, such as chronic diarrhea, abdominal pain, hematochezia, anemia, hypoproteinemia, among other symptoms[3]. Patients with irritable bowel syndrome are more likely to accompany with NLH[4]. During the clinical diagnosis and treatment of patients with terminal ileal NLH, we found that probiotics could improve gastrointestinal symptoms.
NLH may be associated with a risk factor for intestinal lymphoma, as the resolution of gastrointestinal tract nodular lymphoid hyperplasia has been shown following chemotherapy for extraintestinal lymphoma[5]. However, the pathogenesis of NLH has not been fully elucidated yet. A frequently proposed hypothesis implicates an intestinal antigenic trigger, possibly infectious, that leads to the repetitive stimulation and eventual hyperplasia of the lymphoid follicles, which may originate from proliferative plasma cell precursors related to a maturational defect during the development of B lymphocytes[6]. NLH has been reported in patients with human immunodeficiency virus, common variable immunodeficiency, Giardia lamblia infection, helicobacter pylori (H.pylori) infection, familial adenomatous polyposis, and Gardner’s syndrome[7]. The intestinal microbiome plays an essential role in the immune system; gut microbiota that colonize the human intestinal tract form a mutual symbiotic relationship with the host and play a key role in regulating the host immune system and metabolism[8,9]. Conventionally, alterations in the gut microbiota are closely related to inflammatory bowel disease, obesity, colonic adenoma, and colorecatal cancer[10-13], but whether the gut flora plays a role in NLH is unclear.
In this study, we performed bacterial 16S rRNA gene amplicon sequencing of mucosal tissue samples to study the gut microflora dysbiosis that is associated with terminal ileal NLH to determine what alterations occur in microflora and explore the correlation between the intestinal microflora and terminal ileal NLH. Moreover, we used molecular bioinformatic technology to predict the metabolic pathways that are involved in terminal ileal NLH, which will provide the possibility for further targeted intervention therapy.
A total of 30 patients who underwent a colonoscopy in the Digestive Endoscopy Center at Jiading Branch of Shanghai General Hospital (Shanghai, China) from January 2021 to April 2021 were recruited for this study. A total of 15 Patients with terminal ileal NLH (11 males and 4 females aged 24-44 years)were assigned to the test group, while 15 healthy volunteers (7 males and 8 females aged 30-44 years)were assigned to the control group after undergoing a routine physical examination. There were no statistically significant differences in the general data between the groups (P > 0.05). Among the 15 patients with terminal ileal NLH, the most common symptom was diarrhea, followed by abdominal pain and abdominal distension.
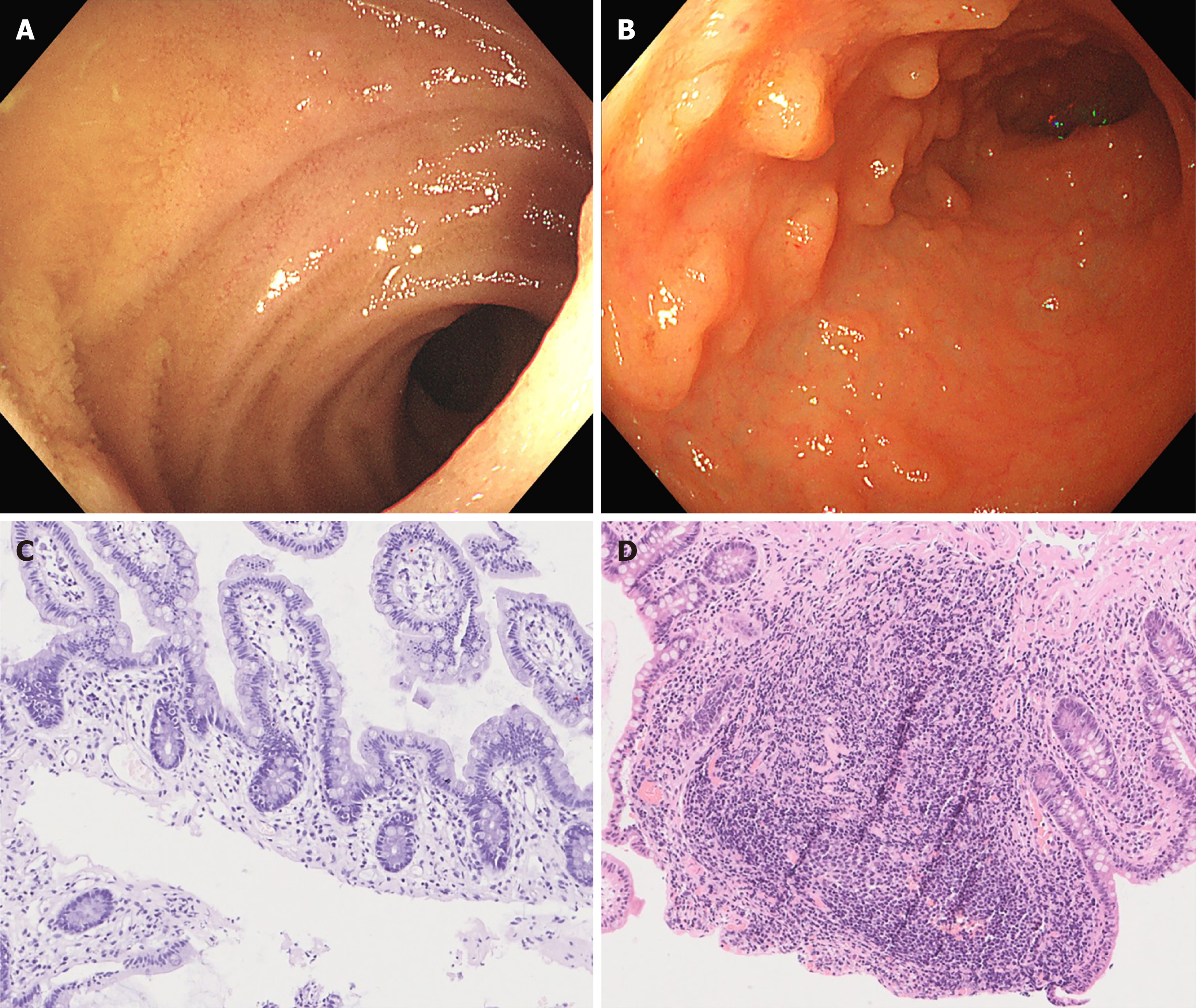
Endoscopic images were reviewed and confirmed by the endoscopic team. Terminal ileal NLH was diagnosed using endoscopy and histopathology (Figure 1). We confirmed that there was no history of antibiotic or probiotic administration within the previous two months for all the subjects. Patients with diabetes mellitus, inflammatory bowel disease, previous colon resection, colorectal cancer, or a body mass index ≥30 kg/m2 were excluded. This research was approved by the research ethics boards of Shanghai General Hospital (2021KY085), and written informed consent was obtained from all the patients before sample collection. We collected mucosal biopsy samples that were obtained via colonoscopy from both groups. All samples were frozen immediately after sampling and stored at -80 °C.
Microbial genomic DNA was extracted from each sample using the E.Z.N.A.® Stool DNA Kit (Omega Bio-tek, Inc., GA) according to the manufacturer’s instructions. The samples were suspended in 790 μl of sterile lysis buffer (4M guanidine thiocyanate; 10% N-lauroyl sarcosine; and 5% N-lauroyl sarcosine-0.1 M phosphate buffer [pH, 8.0]) in a 2-ml screw-cap tube containing 1 gof glass beads (0.1mm BioSpec Products, Inc., United States). This mixture was vortexed vigorously and subsequently incubated at 70°C for 1 h. After incubation by bead beating for 10 min at maximum speed, the extracted DNA was stored at -20 °C for further analysis.
The V3-V4 region of the bacterial 16S ribosomal RNA gene from each sample was amplified using the universal bacterial primers F1 and R2 (5’-CCTACGGGNGGCWGCAG-3’and 5’-GACTACHVGGGTATCTAATCC-3’); these primers correspond to positions 341 to 805 in the Escherichia coli 16S rRNA gene. The PCR reactions were run in a T100™ Thermal Cycler PCR system (Bio-Rad Laboratories, Inc., United States) using the following protocol: 3 min of denaturation at 95 °C, followed by 21 0.5-min denaturation cycles at 94 °C, 0.5 min of annealing at 54 °C, and 0.5 min of elongation at 72 °C, with a final 5 min extension at 72 °C.
The amplicons from different samples were purified using Hieff NGS® DNA Selection Beads (YeasenBiotech Co., Ltd., Shanghai, China). The products were indexed and mixed at equal ratios for sequencing by Shanghai Mobio Biomedical Technology Co., Ltd. using the Miseq platform (Illumina Inc., United States) according to the manufacturer’s instructions.
The raw sequencing data from the 16S rRNA gene V3-V4 regions and the accompanying information are available in the Sequence Read Archive database under accession number PRJNA759383.
Clean data was extracted from the raw data using USEARCH version 11.0.667 (http://www.drive5.com/usearch/). Quality-filtered sequences were clustered into unique sequences and sorted in order of decreasing abundance to identify representative sequences using UPARSE according to the UPARSE Operational Taxonomic Units (OTUs) analysis pipeline, with singletons being omitted. OTUs were classified based on a 97% similarity after the chimeric sequences were removed using UPARSE version 7.1 (http://drive5.com/uparse/), after which they were annotated using the SILVA reference database (SSU138). The number of common OTUs in both groups was calculated and the results were shown using a Venn diagram.
Alpha diversity, which reflects the diversity of microbiome community, was obtained by analyzing the ACE estimator, Chao 1 estimator, Shannon-Wiener diversity index, and Simpson diversity index using Mothur version 1.42.1. The larger the Chao 1 or ACE index, the higher the gut flora abundance, whereas the higher the Shannon or Simpson index, the higher the community diversity.
To visualize the structural diversity of the gut microbiome in the discovery group, we used a principal coordinates analysis (PCoA) and nonmetric multidimensional scaling (NMDS) plots based on the Bray-Curtis distances. The corresponding statistical significance of the beta diversity was measured separately using an Adonis analysis.
To compare the microbial communities at each taxonomic level between the groups, significant between-group differences in the microbial composition were analyzed using a Wilcoxon rank-sum test. A linear discriminant analysis effect size (LEfSe) was used to show the maximum difference in the microbial structures between the groups (LEfSe version 1.1, https://github.com/SegataLab/Lefse) to determine the specific bacterial taxa and predominant bacteria that are related to terminal ileal NLH. The results of the microbiome heatmap analysis, as provided by a random forests model, revealed a discriminatory intestinal microbiome between the two groups.
The Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) 2 version 2.4.1 (https://github.com/picrust/picrust2/wiki) was used to predict different metabolic pathways and orthologous groups between the groups according to the yoto Encyclopedia of Genes and Genomes (KEGG) database.
Non-parametric Mann-Whitney U tests were used to determine if there were significant differences between the groups. A Student’s t-test was performed using SPSS for Windows, version 20.
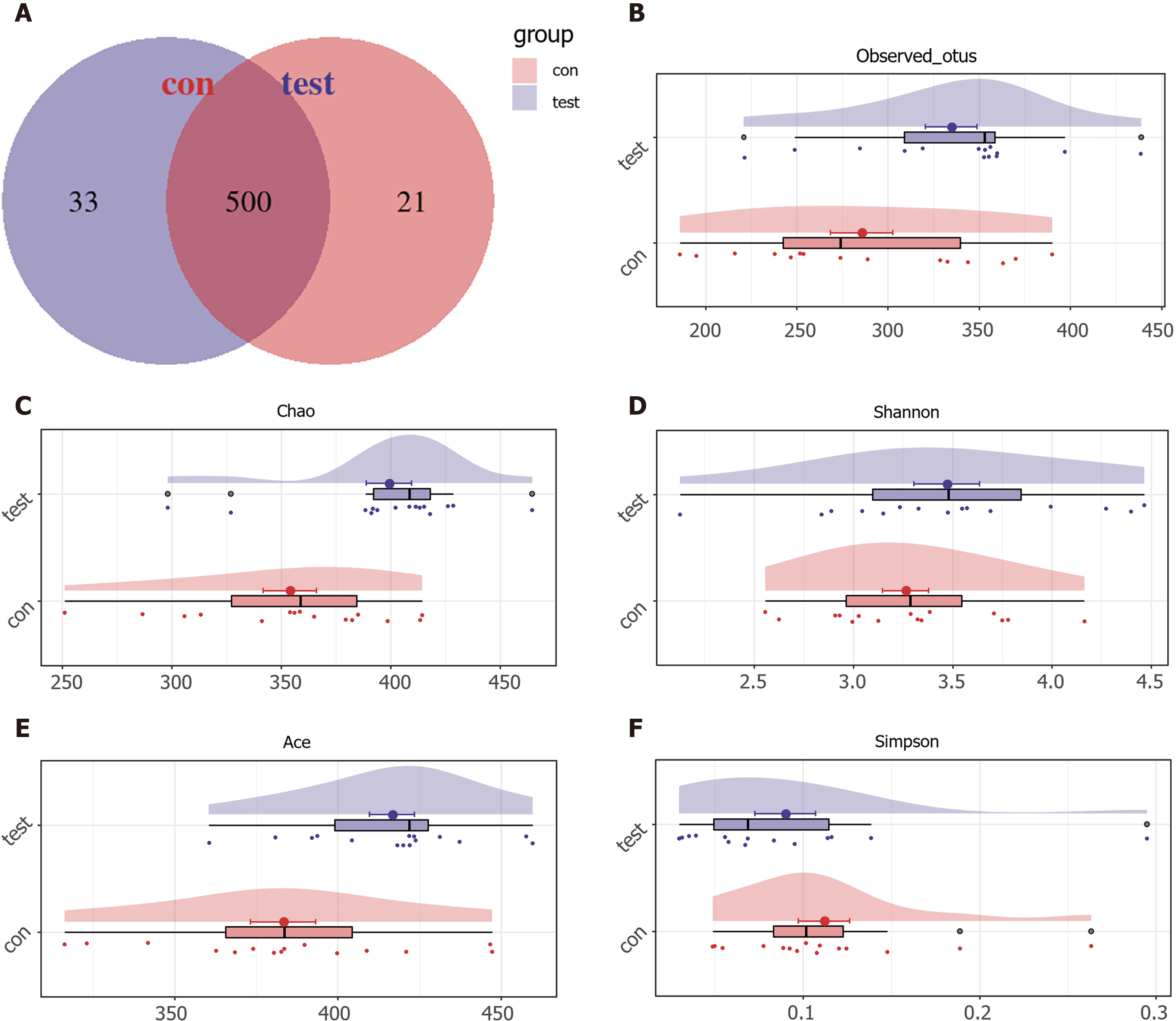
In total, 1530914 usable sequences were obtained from 30 samples using Illumina high-throughput sequencing technology. From these, 1263179 high-quality sequences were selected, with an average of 42106 sequences per sample. Using 97% as the similarity cutoff, we generated 554 OTUs. The Venn diagram showed that 500 of the 554 OTUs were shared by both groups, whereas 33 OTUs were unique to the test group, and 21 were specific to the control group (Figure 2A).
The alpha diversity of the intestinal mucosal microflora was higher for the test group than it was for the control group. As estimated by the observed OTUs in each sample and the ACE and Chao indexes, which reflect the richness of the species diversity, the microbial diversity was significantly higher in the test group than it was in the control group (P < 0.05).The index values for the alpha diversity analysis are shown in Figures 2B-F.
The resulting rarefaction curves indicate that the microbial richness of the sampled guts was near saturation at the applied sequencing depth, which was sufficient to identify most of the bacterial community members in each individual. The Shannon-Wiener curve based on the OTUs was already flat, indicating that our sequencing depth was already adequate. The specaccum species accumulation curves revealed that the OTU richness approached saturation in all the samples (Supplemen
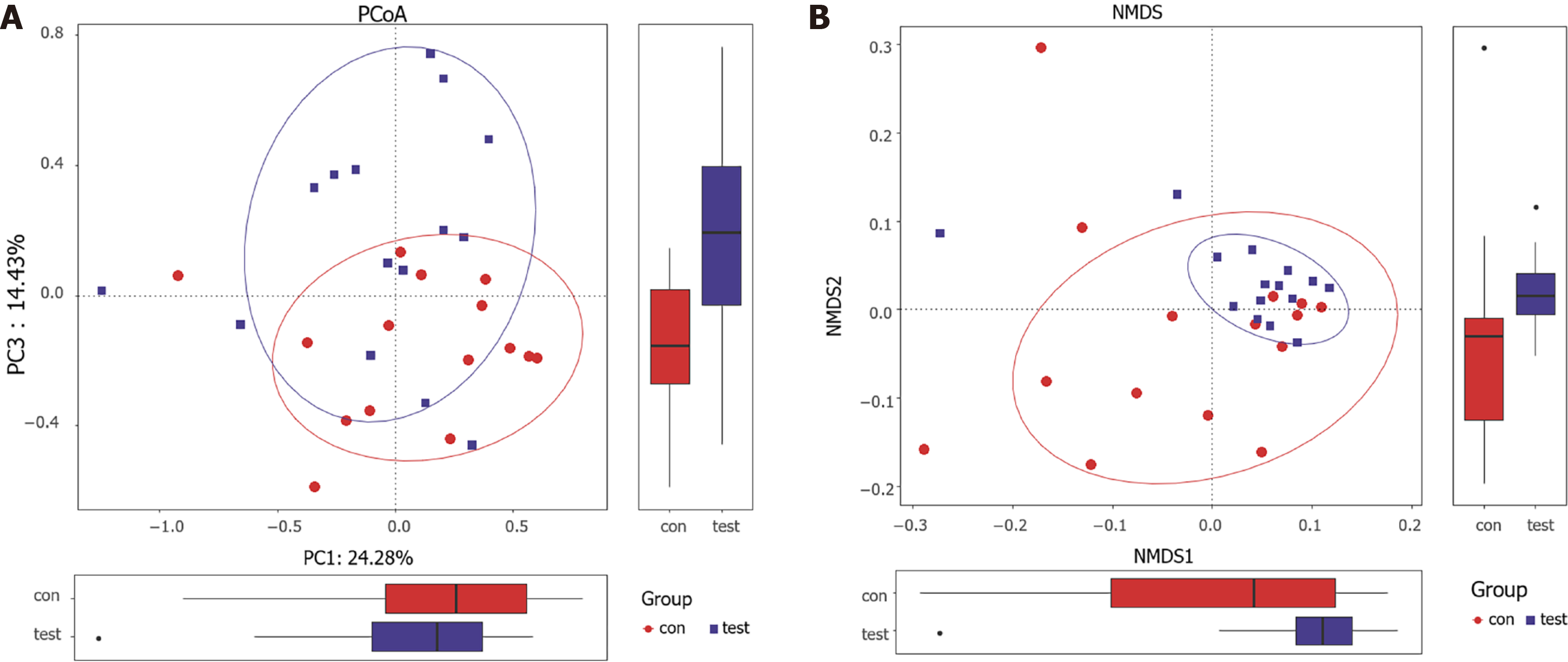
To evaluate the similarities between all samples, the ecologic distances, which were calculated based on the Bray-Curtis distances, were visualized using a PCoA plot. A certain tendency of separation was found between both groups, indicating that the bacterial flora differences in the overall structure of gut microbiota existed between the groups (Figure 3A). Moreover, a non-metric multidimensional scaling analysis based on the Bray-Curtis distances showed a significant difference in gut microbiomes between both groups (Figure 3B).
Additionally, an Adonis analysis showed that there was a significant difference between the two groups (P < 0.05). Based on the unweighted and weighted UniFrac distances, a PCoA also showed that the microbial composition of the test group deviated from the control group (Adonis: P = 0.0142 and P = 0.0467, respectively).
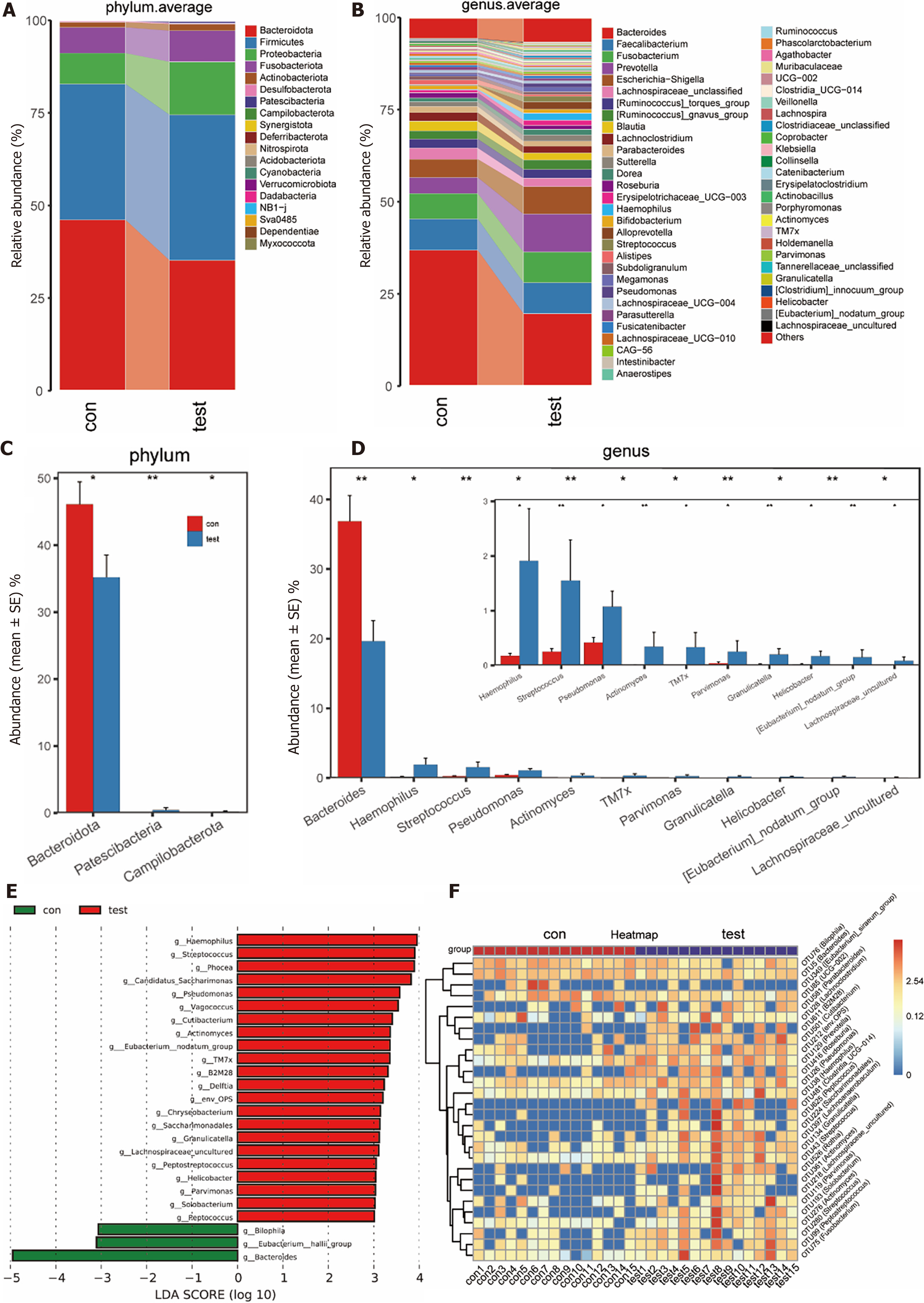
A total of 19 phyla were detected by classifying the species of all OTUs in the terminal ileal mucosa. At the phylum level, the gut microbiota of both groups was dominated by Bacteroidetes and Firmicutes, followed by (on average) Proteobacteria. The proportions of dominant flora in the test group were 35.19%, 39.29%, and 14.31% respectively, while those of the control group were 46.11%, 36.68%, and 8.34%, respectively. The average relative abundance of the microbiome at the phylum level is shown in Figure 4A.
At the genus level, the gut microbiota was dominated by Bacteroides in the control group, followed by Faecalibacterium, Fusobacterium, Escherichia-Shigella, and Prevotella with proportions of 36.85%, 8.47%, 6.93%, 4.96%, and 4.33%, respectively. Correspondingly, Bacteroides was the most dominant bacteria in the test group, followed by Prevotella,Faecalibacterium, Fusobacterium, and Escherichia−Shigella, with proportions of 19.63%, 10.31%, 8.39%, 8.33%, and 7.43%, respectively. The average relative abundance of the microbiome at the genus level is shown in Figure 4B.
There were significant differences in the microbial composition between the groups, as analyzed by the Wilcoxon rank-sum test. At the phylum level, Bacteroidetes were significantly lower in the test group than in the control group. In contrast, Patescibacteria and Campilobacterota were significantly higher in the test group than in the control group. At the genus level, the number of Bacteroides was significantly lower in the test group than in the control group. Conversely, the abundances of Haemophilus, Streptococcus, Pseudomonas, Actinomyces, TM7X, Parvimonas, Granulicatella, Helicobacter and the [Eubacterium] nodatum group, among others, were significantly higher in the test group than in the control group (Figures 4C and D).
A LEfSe was used to show the maximum difference in microbial structures between the groups to determine the specific bacterial taxa and predominant bacteria in the patients with terminal ileal NLH. This analysis showed that the abundances of various genera, including Haemophilus, Streptococcus, Phocea, Candidatus_saccharimonas, Pseudomonas, Vagococcus, Cutibacterium, Actinomyces, the Eubacterium nodatum group, TM7X, Delftia, Chryseobacterium, Peptostreptococcus, Helicobacter, Parvimonas, Solobacterium, and Peptococcus, among others, were significantly higher in the test group than in the control group. Conversely, the abundances of Bacteroides, Bilophila, and the Eubacterium hallii group were significantly higher in the control group than in the test group (Figure 4E).
The results of the heatmap analysis of the microbiomes using a random forest model revealed a discriminatory intestinal microbiome between both groups. A total of 28 OTUs were found to be different between the sample groups. Among these OTUs, 24 were more abundant in the test group than in the control group; these OTUs belonged to the genera of Rothia, Roseburia, Cutibacterium, Peptococcus, Parabacteroides, Lachnoanaerobaculum, Actinomyces, Streptococcus and SaccharimonAdales, Solobacterium, Peptostreptococcus, Granulicatella, Parvimonas, Lachnoclostridium, Pseudomonas, Fusobacterium, Haemophilus, Prevotella, and Clostridia UCG-014. Bacteroides, Bilophila, and the [Eubacterium] Siraeum group were more abundant in the control group than in the test group (Figure 4F).
PICRUSt2 version 2.4.1 was used to predict metabolic pathways and orthologous groups according to the KEGG database, and a LEfSe was subsequently used to sort the different metabolic pathways and orthologous groups between the two groups. The results demonstrated that Photosynthesis, D Alanine metabolism, C5 Branched dibasic acid metabolism, Peptidoglycan biosynthesis, Aminoacyl tRNA biosynthesis, Bacterial chemotaxis, ABC transporters, D glutamine and D glutamate metabolism, synthesis and degradation of ketone bodies, ulfur relay system, ribosome, mismatch repair, homologous recombination, Glycerophospholipid metabolism, base excision repair, DNA replication, butanoate metabolism, bacterial secretion system, terpenoid backbone biosynthesis, and styrene degradation were significantly higher in test group compared to the control group. However, glycosaminoglycan degradation, secondary bile acid biosynthesis, sphingolipid metabolism, biotin metabolism, pentose and glucuronate interconversions, galactose metabolism, streptomycin biosynthesis, cyanoamino acid metabolism, and alanine aspartate and glutamate metabolism pathways were all significantly higher in the control group than in the test group. Altered metabolic pathways, orthologous groups, and modules in both groups are presented in Figure 5.
Terminal ileal NLH is commonly detected through colonoscopy. The pathogenesis of terminal ileal NLH remains unclear, it is generally regarded that infection-induced immune responses play a key role. Alterations of the gut microbiota are closely related to immune-related diseases. However, whether the gut flora plays a role in terminal ileal NLH is unclear. Currently, there are no studies that have reported on the relationship between the intestinal flora and terminal ileal NLH. In this study, alpha diversity was higher in the test group than in the control group. Helicobacter, Fusobacterium nucleatum, Actinomyces, TM7X, and Peptostreptococcus were significantly more abundant in the test group than in the control group. Additionally, the Peptidoglycan and Aminoacyl tRNA biosynthesis pathways were significantly more abundant in the test group than in the control group.
In this study, we found that the bacterial diversity was significantly higher in the terminal ileal NLH group than in the control group, suggesting the presence of a small intestinal bacterial overgrowth (SIBO). SIBO is defined as bacterial overgrowth in the small intestine caused by an abnormally high number of bacteria and/or changes in the kinds of bacteria; it is accompanied by an overgrowth of bacteria in the small bowel in excess of 105 colony forming units per milliliter in upper gut aspirate culture[14]. This is due to the bacteria migrating into the small intestine from distal intestinal tract, resulting in intestinal mucosal inflammation and permeability and villi damage, which mainly manifests as nutrient malabsorption, abdominal pain and distension, diarrhea, and intestinal motility abnormity[15]. SIBO is closely associated with many diseases, including colorectal cancer, irritable bowel syndrome, inflammatory bowel disease, and non-alcoholic fatty liver disease[16-19]. In this study, we found an increased alpha diversity of intestinal flora in the test group than in the control group. This observation may be related to the local inflammatory response caused by the overgrowth of intestinal bacteria, which results in NLH.
Bacteroidetes and Firmicutes make up most of the human intestinal flora, with a higher abundance of Bacteroidetes. In this study, Bacteroidetes and Firmicutes were the dominant bacteria in both groups, followed by Proteobacteria. Similarly, Bacteroides was the most dominant bacteria in both groups at the genus level. However, Bacteroidetes and Bacteroides were significantly less abundant in the test group than in the control groups, suggesting that Bacteroides may play a protective role in the development of terminal ileal inflammation and NLH.
In our research, H.pylori was higher in abundance in the test group. H.pylori, which is a proteobacteria, is considered to be a carcinogenic factor of gastric cancer. Gastric mucosa-associated lymphoid tissue (MALT) lymphoma is related to H.pylori[20,21], as most patients with MALT can achieve long-term clinical remission after H.pylori eradication[22,23]. H.pylori is thought to be an antigenic stimulant that can activate the NF-κB pathway[24] and induce pro-inflammatory cytokines expression [25,26]. Persistent inflammation promotes the formation of mucosal lymphoid follicles, typically consisting of B lymphocytes, which might contribute to the genesis of gastric MALT lymphoma once the inflammatory courses are uncontrolled[27]. Khuroo et al studied a large cohort of patients (n = 40) with NLH that was etiologically related to H.pylori infection. Compared with patients with consistent H.pylori infection, patients with eradicated H.pylori showed a significant clinical response and lesion regression/resolution[28]. However, the location was limited to the postbulbar duodenum (second and third parts) and duodenojejunal junction in these cases. In our study, the H.pylori abundance increased in test group. Moreover, it has been reported that NLH may be associated with an increased risk of parenteral lymphoma. However, currently, there are no relevant reports on the correlation and causal relationship between terminal ileal NLH and H.pylori, which is worthy of studying. When treating patients with H.pylori, we suggest that endoscopists should routinely observe the terminal ileum.
In this study, the abundance of Actinomycetes and TM7x increased in the test group. Actinomycetes mostly reside in the oral cavity, upper respiratory tract, digestive tract, and urogenital tract in humans and animals, as part of the normal flora. Actinomycetes are recognized to be a cause of chronic appendicitis. Actinomycetes are also associated with Crohn-like appendicitis with significant fibrosis, transmural inflammation, lymphoid hyperplasia, and granuloma[29]. TM7x is a saccharifying bacteria, which is a parasitic bacterium that interacts with core members of Actinomycetes. A previous study suggested that ultra-small bacteria may have the ability to regulate the immune response of normal hosts, and there was a signal overlap between TM7x and basibiont Actinomyces odontolyticus species (XH001) by metabolic pathway prediction[30]. Moreover, through in vitro experiments, the authors demonstrated that TM7x inhibited TNF-α expression in XH001-induced macrophages[31]. Consequently, the interaction between Actinomycetes and TM7x may promote terminal ileal NLH.
In this study, we found that the abundance of Fusobacterium nucleatum increased in the test group, and this increase is possibly related to terminal ileal NLH. Fusobacterium nucleatum has been reported to be enriched in colorectal cancer tissues and played a crucial role in the occurrence and development of colorectal cancer[32]. It can adhere to and invade intestinal epithelial cells and activate the β-catenin pathway by releasing FadA adhesin and binding with cadherin E-cadherin, thus promoting inflammation and tumor response[33]. Recently, it has been reported that Fusobacterium nucleatum macromolecules (> 50 KDA) have a proinflammatory effect on human intestinal epithelium, and the outer membrane vesicles can promote the secretion of proinflammatory cytokines, including IL-8 and TNFα by epithelial cells[34]. Further animal experiments also verified the proinflammatory effect of Fusobacterium nucleatum on intestinal epithelium. Thus, Fusobacterium nucleatum may be associated with the development of terminal NLH.
In our study, the abundance of Peptostreptococcus increased in the test group. Anaerobic peptostreptococcus and Peptostreptococcus magnus are most commonly in the genus of Peptostreptococcus, among which Anaerobic peptostreptococcus is the most common pathogen. Anaerobic peptostreptococcus is a gram-positive anaerobic bacteria commonly residing in the oral cavity and digestive tract. The abundance of Anaerobic peptostreptococcus in the stool samples of patients with colorectal cancer was reported to be higher than healthy volunteers. In vitro studies have shown that Anaerobic digestion streptococcus interacts with TLR2 and TLR4 in colon cells and increases the level of active oxides, thereby promoting cholesterol synthesis and cell proliferation[35]. The PCWBR2 integrin α2/β1-PI3K-Akt-NF-κB signal axis has been reported to be involved in the development of colorectal cancer[36]. Peptostreptococcus may be related to terminal ileal inflammation and NLH, although further confirmation of this is needed in the future.
Metabolic pathways, such as the Peptidoglycan biosynthesis and Aminoacyl tRNA biosynthesis, increased in the test group in the current study. Peptidoglycan, a bacterial cell wall component, is a conserved pathogen-associated molecule that is involved in the innate immune system because it recognizes pattern recognition receptors that are secreted and expressed in or on the cell surface[37]. Transfer RNAs (tRNAs) mainly participates in protein translation by transporting amino acids to the ribosome. Nevertheless, accumulating evidence has shown that tRNAs are closely associated with various physiological and pathological processes such as immune regulation. Aminoacyl-tRNA synthetases (ARSs) are essential components of translation in all living species, and it has taken scientists decades to confirm that eukaryotic ARSs act as global cell signaling mediators to regulate cell homeostasis beyond their intrinsic function as protein synthesis enzymes. Recent discoveries have revealed that ubiquitously expressed standby cytoplasmic ARSs sense and respond to danger signals and regulate immunity against infections, indicating their potential as therapeutic targets for infectious diseases[38,39]. Perhaps Peptidoglycan biosynthesis and Aminoacyl-tRNA biosynthesis might act as the targeted intervention sites.
The etiology of terminal ileal NLH has not been fully elucidated yet. In this study, we firstly analyzed the diversity and composition of intestinal flora in the mucosal tissues of the patients with terminal ileal NLH and predicted the metabolic pathways using 16S-rRNA technology. We subsequently found that terminal ileal NLH was related to the disturbance of the intestinal flora and certain microflora might act on the small intestinal mucosa thereby causing terminal ileal NLH. Moreover, the metabolic pathways that were predicted using PICRUSTs are possibly involved in terminal ileal NLH, which provides novel ideas for further exploration of potential molecular mechanisms. Diarrhea was frequently commonly found in patients with terminal ileal NLH, and some patients may get a satisfactory effect through probiotic supplementation. Therefore, exploring intestinal flora changes, seeking related bacteria genera in patients with terminal ileal NLH, and supplementing targeted protective bacteria or clearing targeted bacteria may reduce the risk of lymphoma and improve patient symptoms.
This study has some limitations. First, the sample size was relatively small. Although our preliminary results reveal that there was a significant difference between the two groups, studies with larger sample sizes covering different regions and populations are necessary to confirm the findings. Second, the male-female ratio in the NLH group was not balanced in this study due to the nature of terminal ileal NLH, which is thought to be significantly more common in men than women. Although the incidence of terminal ileal NLH in both men and women has not been investigated through a large-scale study, Lin et al observed that males outnumbered females by approximately four to one in a small-scale study[3]; their results support, to a certain degree, the suggestion that there is a higher frequence of terminal ileal NLH in males than females. We also performed a correlation analysis to compare the intestinal flora with gender using the Multivariable Association with Linear Models2, and we found that there was no correlation between the intestinal floras and gender at the phylum, genus, or OTU levels. To obtain more rigorous results, we will perform a large-scale study and ensure that there is an equal gender ratio among groups. Third, this study was only a preliminary correlation analysis between the intestinal flora and terminal ileal NLH, and no further research on the related mechanisms was performed. Further studies using animal testing in vivo and in vitro cellular experiments can be developed once our findings are verified in larger populations.
Intestinal flora disturbances are related to terminal ileal NLH, and our results show that certain microflora may act on the small intestinal mucosa and cause terminal ileal NLH. Maintaining the intestinal flora balance and supplementing targeted protective bacteria could improve terminal ileal NLH symptoms and potentially reduce the risk of lymphoma transformation.
Nodular lymphoid hyperplasia (NLH) in the small intestine is a rare benign lesion characterized by multiple small nodules on the intestinal surface. NLH is linked to the immune system, and it may result from accumulation of plasma-cell precursors due to a maturational defect during the development of B lymphocytes. The intestinal microbiome plays an essential role in the immune system. However, whether the gut flora plays a role in NLH is unclear.
To explore the correlation between intestinal flora and terminal ileal NLH and predict the metabolic pathways that are involved in terminal ileal NLH.
To investigate the characteristics of the mucosal microbiata in patients with terminal ileal NLH for seeking related bacteria genera and bringing a new idea for related mechanisms.
A total of 30 patients who underwent a colonoscopy were recruited for this study. A total of 15 Patients with terminal ileal NLH were assigned to the test group, while 15 healthy volunteers were assigned to the control group after undergoing a routine physical examination. We collected mucosal biopsy samples that were obtained via colonoscopy from both groups. We subsequently performed 16S-rRNA gene amplicon sequencing of these samples, and the results were evaluated using alpha diversity, beta diversity and microbial composition analyses. The Phylogenetic Investigation of Communities by Reconstruction of Unobserved States was used to predict the metabolic pathways and orthologous groups according to the Kyoto Encyclopedia of Genes and Genomes database.
The terminal ileal NLH group showed an increased alpha diversity.The overall intestinal microbiota in the NLH group was significantly different from that of the control group. The relative abundance of phylum Bacteroidetes was significantly lower in the NLH group, while that of Patescibacteria and Campilobacterota was significantly higher. The abundance of the genus Bacteroides was significantly lower in the test group. Conversely, the relative abundances of Haemophilus, Streptococcus, Pseudomonas, Actinomyces, TM7X, Fusobacterium nucleatum, Parvimonas, Granulicatella, Helicobacter, and the [Eubacterium] nodatum group were significantly higher in the test group. Metabolic pathways such as Peptidoglycan biosynthesis and Aminoacyl tRNA biosynthesis were both increased in the test group.
Maintaining the microbial balance and supplementing targeted protective bacteria could improve symptoms and potentially reduce the risk of lymphoma transformation in patients with terminal ileal NLH.
Further research on the related mechanisms was needed to be performed in future. Further studies using animal testing in vivo and in vitro cellular experiments can be developed once our findings are verified in larger populations.
The authors would like to acknowledge Chen H for skillful technical assistance.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Fujimori S, Srivastava D S-Editor: Wang LL L-Editor: A P-Editor: Yu HG
| 1. | Ranchod M, Lewin KJ, Dorfman RF. Lymphoid hyperplasia of the gastrointestinal tract. A study of 26 cases and review of the literature. Am J Surg Pathol. 1978;2:383-400. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 97] [Cited by in F6Publishing: 100] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 2. | Rambaud JC, De Saint-Louvent P, Marti R, Galian A, Mason DY, Wassef M, Licht H, Valleur P, Bernier JJ. Diffuse follicular lymphoid hyperplasia of the small intestine without primary immunoglobulin deficiency. Am J Med. 1982;73:125-132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 3. | Lin R, Lu H, Zhou G, Wei Q, Liu Z. Clinicopathological and Ileocolonoscopic Characteristics in Patients with Nodular Lymphoid Hyperplasia in the Terminal Ileum. Int J Med Sci. 2017;14:750-757. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 4. | Piscaglia AC, Laterza L, Cesario V, Gerardi V, Landi R, Lopetuso LR, Calò G, Fabbretti G, Brisigotti M, Stefanelli ML, Gasbarrini A. Nodular lymphoid hyperplasia: A marker of low-grade inflammation in irritable bowel syndrome? World J Gastroenterol. 2016;22:10198-10209. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 11] [Cited by in F6Publishing: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 5. | Jonsson OT, Birgisson S, Reykdal S. Resolution of nodular lymphoid hyperplasia of the gastrointestinal tract following chemotherapy for extraintestinal lymphoma. Dig Dis Sci. 2002;47:2463-2465. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 11] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 6. | Hermans PE, Huizenga KA, Hoffman HN, Brown AL Jr, Markowitz H. Dysgammaglobulinemia associated with nodular lymphoid hyperplasia of the small intestine. Am J Med. 1966;40:78-89. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 7. | Albuquerque A. Nodular lymphoid hyperplasia in the gastrointestinal tract in adult patients: A review. World J Gastrointest Endosc. 2014;6:534-540. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 58] [Cited by in F6Publishing: 48] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 8. | Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30:492-506. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 789] [Cited by in F6Publishing: 1415] [Article Influence: 353.8] [Reference Citation Analysis (0)] |
| 9. | Bouskra D, Brézillon C, Bérard M, Werts C, Varona R, Boneca IG, Eberl G. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507-510. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 771] [Cited by in F6Publishing: 754] [Article Influence: 47.1] [Reference Citation Analysis (0)] |
| 10. | Sheehan D, Shanahan F. The Gut Microbiota in Inflammatory Bowel Disease. Gastroenterol Clin North Am. 2017;46:143-154. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 11. | Rosenbaum M, Knight R, Leibel RL. The gut microbiota in human energy homeostasis and obesity. Trends Endocrinol Metab. 2015;26:493-501. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 299] [Cited by in F6Publishing: 283] [Article Influence: 31.4] [Reference Citation Analysis (0)] |
| 12. | Peters BA, Dominianni C, Shapiro JA, Church TR, Wu J, Miller G, Yuen E, Freiman H, Lustbader I, Salik J, Friedlander C, Hayes RB, Ahn J. The gut microbiota in conventional and serrated precursors of colorectal cancer. Microbiome. 2016;4:69. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 136] [Cited by in F6Publishing: 149] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 13. | Tilg H, Adolph TE, Gerner RR, Moschen AR. The Intestinal Microbiota in Colorectal Cancer. Cancer Cell. 2018;33:954-964. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 398] [Cited by in F6Publishing: 466] [Article Influence: 77.7] [Reference Citation Analysis (0)] |
| 14. | Bouhnik Y, Alain S, Attar A, Flourié B, Raskine L, Sanson-Le Pors MJ, Rambaud JC. Bacterial populations contaminating the upper gut in patients with small intestinal bacterial overgrowth syndrome. Am J Gastroenterol. 1999;94:1327-1331. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in F6Publishing: 113] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 15. | Bures J, Cyrany J, Kohoutova D, Förstl M, Rejchrt S, Kvetina J, Vorisek V, Kopacova M. Small intestinal bacterial overgrowth syndrome. World J Gastroenterol. 2010;16:2978-2990. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 383] [Cited by in F6Publishing: 332] [Article Influence: 23.7] [Reference Citation Analysis (3)] |
| 16. | Liang S, Xu L, Zhang D, Wu Z. Effect of probiotics on small intestinal bacterial overgrowth in patients with gastric and colorectal cancer. Turk J Gastroenterol. 2016;27:227-232. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 17. | Ghoshal UC, Srivastava D. Irritable bowel syndrome and small intestinal bacterial overgrowth: meaningful association or unnecessary hype. World J Gastroenterol. 2014;20:2482-2491. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 72] [Cited by in F6Publishing: 64] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 18. | Ricci JERJ, Chebli LA, Ribeiro T, Castro ACS, Gaburri PD, Pace FHDL, Barbosa KVBD, Ferreira LEVVDC, Passos MDCF, Malaguti C, Delgado ÁHDA, Campos JD, Coelho AR, Chebli JMF. Small-Intestinal Bacterial Overgrowth is Associated With Concurrent Intestinal Inflammation But Not With Systemic Inflammation in Crohn’s Disease Patients. J Clin Gastroenterol. 2018;52:530-536. [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 22] [Article Influence: 3.7] [Reference Citation Analysis (1)] |
| 19. | Fialho A, Fialho A, Thota P, McCullough AJ, Shen Bo. Small Intestinal Bacterial Overgrowth Is Associated with Non-Alcoholic Fatty Liver Diseasel. J Gastrointestin Liver Dis. 2016;25:159-165. [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 48] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 20. | Wotherspoon AC, Ortiz-Hidalgo C, Falzon MR, Isaacson PG. Helicobacter pylori-associated gastritis and primary B-cell gastric lymphoma. Lancet. 1991;338:1175-1176. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 4] [Reference Citation Analysis (0)] |
| 21. | Ferrero RL, Avé P, Radcliff FJ, Labigne A, Huerre MR. Outbred mice with long-term Helicobacter felis infection develop both gastric lymphoid tissue and glandular hyperplastic lesions. J Pathol. 2000;191:333-340. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 5] [Reference Citation Analysis (0)] |
| 22. | Bayerdörffer E, Neubauer A, Rudolph B, Thiede C, Lehn N, Eidt S, Stolte M. Regression of primary gastric lymphoma of mucosa-associated lymphoid tissue type after cure of Helicobacter pylori infection. MALT Lymphoma Study Group. Lancet. 1995;345:1591-1594. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 460] [Reference Citation Analysis (1)] |
| 23. | Chen LT, Lin JT, Tai JJ, Chen GH, Yeh HZ, Yang SS, Wang HP, Kuo SH, Sheu BS, Jan CM, Wang WM, Wang TE, Wu CW, Chen CL, Su IJ, Whang-Peng J, Cheng AL. Long-term results of anti-Helicobacter pylori therapy in early-stage gastric high-grade transformed MALT lymphoma. J Natl Cancer Inst. 2005;97:1345-1353. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 154] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 24. | Ferrero RL, Avé P, Ndiaye D, Bambou JC, Huerre MR, Philpott DJ, Mémet S. NF-kappaB activation during acute Helicobacter pylori infection in mice. Infect Immun. 2008;76:551-561. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 35] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 25. | Kuo SH, Yeh PY, Chen LT, Wu MS, Lin CW, Yeh KH, Tzeng YS, Chen JY, Hsu PN, Lin JT, Cheng AL. Overexpression of B cell-activating factor of TNF family (BAFF) is associated with Helicobacter pylori-independent growth of gastric diffuse large B-cell lymphoma with histologic evidence of MALT lymphoma. Blood. 2008;112:2927-2934. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 44] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 26. | Munari F, Fassan M, Capitani N, Codolo G, Vila-Caballer M, Pizzi M, Rugge M, Della Bella C, Troilo A, D'Elios S, Baldari CT, D'Elios MM, de Bernard M. Cytokine BAFF released by Helicobacter pylori-infected macrophages triggers the Th17 response in human chronic gastritis. J Immunol. 2014;193:5584-5594. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 43] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 27. | Sagaert X, Van Cutsem E, De Hertogh G, Geboes K, Tousseyn T. Gastric MALT lymphoma: a model of chronic inflammation-induced tumor development. Nat Rev Gastroenterol Hepatol. 2010;7:336-346. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 95] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 28. | Khuroo MS, Khuroo NS, Khuroo MS. Diffuse duodenal nodular lymphoid hyperplasia: a large cohort of patients etiologically related to Helicobacter pylori infection. BMC Gastroenterol. 2011;11:36. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 29. | Horvath BA, Maryamchik E, Miller GC, Brown IS, Setia N, Mattia AR, Lamps L, Lauwers GY, Rosenberg E, Misdraji J. Actinomyces in Crohn's-like appendicitis. Histopathology. 2019;75:486-495. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 30. | Bor B, Poweleit N, Bois JS, Cen L, Bedree JK, Zhou ZH, Gunsalus RP, Lux R, McLean JS, He X, Shi W. Phenotypic and Physiological Characterization of the Epibiotic Interaction Between TM7x and Its Basibiont Actinomyces. Microb Ecol. 2016;71:243-255. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 31. | He X, McLean JS, Edlund A, Yooseph S, Hall AP, Liu SY, Dorrestein PC, Esquenazi E, Hunter RC, Cheng G, Nelson KE, Lux R, Shi W. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc Natl Acad Sci U S A. 2015;112:244-249. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 311] [Cited by in F6Publishing: 293] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 32. | Liu W, Zhang X, Xu H, Li S, Lau HC, Chen Q, Zhang B, Zhao L, Chen H, Sung JJ, Yu J. Microbial Community Heterogeneity Within Colorectal Neoplasia and its Correlation With Colorectal Carcinogenesis. Gastroenterology. 2021;160:2395-2408. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 64] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 33. | Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195-206. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1511] [Cited by in F6Publishing: 1367] [Article Influence: 124.3] [Reference Citation Analysis (0)] |
| 34. | Engevik MA, Danhof HA, Ruan W, Engevik AC, Chang-Graham AL, Engevik KA, Shi Z, Zhao Y, Brand CK, Krystofiak ES, Venable S, Liu X, Hirschi KD, Hyser JM, Spinler JK, Britton RA, Versalovic J. Fusobacterium nucleatum Secretes Outer Membrane Vesicles and Promotes Intestinal Inflammation. mBio. 2021;12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 45] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 35. | Tsoi H, Chu ESH, Zhang X, Sheng J, Nakatsu G, Ng SC, Chan AWH, Chan FKL, Sung JJY, Yu J. Peptostreptococcus anaerobius Induces Intracellular Cholesterol Biosynthesis in Colon Cells to Induce Proliferation and Causes Dysplasia in Mice. Gastroenterology. 2017;152:1419-1433.e5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 203] [Cited by in F6Publishing: 237] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 36. | Long X, Wong CC, Tong L, Chu ESH, Ho Szeto C, Go MYY, Coker OO, Chan AWH, Chan FKL, Sung JJY, Yu J. Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nat Microbiol. 2019;4:2319-2330. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 124] [Cited by in F6Publishing: 229] [Article Influence: 45.8] [Reference Citation Analysis (0)] |
| 37. | Crump GM, Zhou J, Mashayekh S, Grimes CL. Revisiting peptidoglycan sensing: interactions with host immunity and beyond. Chem Commun (Camb). 2020;56:13313-13322. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 38. | Nie A, Sun B, Fu Z, Yu D. Roles of aminoacyl-tRNA synthetases in immune regulation and immune diseases. Cell Death Dis. 2019;10:901. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 49] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 39. | Park SG, Schimmel P, Kim S. Aminoacyl tRNA synthetases and their connections to disease. Proc Natl Acad Sci. 105:11043-11049. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 262] [Cited by in F6Publishing: 264] [Article Influence: 16.5] [Reference Citation Analysis (0)] |